rm(list = ls())
# Set global R options
options(scipen = 4)
# Set the graphical theme
ggplot2::theme_set(ggplot2::theme_light())
# Set global knitr chunk options
knitr::opts_chunk$set(
warning = FALSE,
message = FALSE,
cache = TRUE
)
set.seed(123)
library(truncnorm)
library(pracma)
library(ggplot2)Quantile Importance Sampling
Multivariate t
Multivariate \(t\) Example
As an example of a higher-dimensional integral, we look at a multivariate \(t\) likelihood and a multivariate Gaussian prior with dimension \(d = 50\), following Polson and Scott (2014). The target integral is:
\[ Z = \int_{\mathbb{R}^d} (1 + \frac{x^T x}{\nu})^{-\frac{\nu + d}{2}} (\frac{\tau}{2\pi})^{d/2} \exp \{-\tau x^T x / 2\} d x. \]
This integral can be written in terms of Kummer’s confluent hypergeometric function of the second kind: \(Z = s^{a}U(a, b, s)\), where \(a = (\nu + d)/2\), \(b = \nu/2 + 1\), \(s = \nu \tau/2\). For the specific values used here: \(d = 50, \tau =1\) and \(\nu = 2\), we can get: \(Z = U(26, 2, 1) = 1.95\times 10^{-29}\).
We show the R codes for comparing QIS with naive Monte Carlo.
Estimation by QIS, and Naive MC
We define three essential functions below:
vertical.gridwill generate an \(x\)-grid for integration using either the exponential weights (for original nested sampling), or uniform for Yakowitz (Quantile Importance Sampling. )sQis simply a sample quantile calculator.
# Generate grid
vertical.grid = function(l,N,type = NULL){
# "u" - uniform
# "e" - exponential
if(type == "u"){
ugrid = runif(N)
res = c(sort(ugrid),1)
# res = sort(runif(N))
}else if(type == "e"){
res = exp(-(0:l)/N)
}
return(res)
}
# Quantile
sQ = function(q,Y){
# q-quantile of Y
N = length(Y)
res = Y[ceiling(N*q)]
return(res)
}Naive Monte Carlo
# Test example
# Prior and Likelihood
set.seed(90210)
d = 50
tau = 1
nu = 2
trueZ <- 1.9445572*10^(-29) ## U(26,2,1), d = 50, nu = 2, s = 1
dtmvr <- function(x, nu){
d = length(x)
logden = -0.5*(nu + d)*log(1 + (t(x)%*%x)/nu)
return(exp(logden))
}
library(LaplacesDemon)
r = 100
mc.naive = NULL
verbose = TRUE
for(i in 1:r){
if(isTRUE(verbose) && i %% 10 == 0)
cat("Iteration ",i, "\n")
M = 10000
# X = rmvn(M, rep(0,d), eye(d))
Y = numeric(M)
for(j in 1:M){
Y[j] = dtmvr(x = rnorm(d, 0, 1), nu = 2)
}
mc.naive = c(mc.naive,mean(Y))
}Iteration 10
Iteration 20
Iteration 30
Iteration 40
Iteration 50
Iteration 60
Iteration 70
Iteration 80
Iteration 90
Iteration 100 mean(mc.naive)[1] 4.538692e-31summary(mc.naive) Min. 1st Qu. Median Mean 3rd Qu. Max.
7.554e-33 3.820e-32 7.599e-32 4.539e-31 1.793e-31 2.362e-29 Quantile Importance Sampling
set.seed(90210)
## QIS
N = 20
# r = 100
mc.qis = NULL
verbose = TRUE
simu.grid.unif = vertical.grid(l=NULL,N,type = "u")
for(i in 1:r){
if(isTRUE(verbose) && i %% 10 == 0)
cat("Iteration ",i, "\n")
M = 10000
# X = rmvn(M,rep(0,d), eye(d))
Y = numeric(M)
for(j in 1:M){
Y[j] = dtmvr(x = rnorm(d, 0, 1), nu = 2)
}
Y = sort(Y)
Lambda = sQ(simu.grid.unif,Y)
x = simu.grid.unif
y = Lambda
# Use a correction term at the boundary: -h^2/12*(f'(b)-f'(a))
# h <- x[2] - x[1]
# ca <- (y[2]-y[1]) / h
# cb <- (y[N]-y[N-1]) / h
# YakoMC <- trapz(x, y) - h^2/12 * (cb - ca)
YakoMC <- trapz(x, y)
# mc.qis = c(mc.qis, trapz(simu.grid.unif,Lambda)) ## QIS Original
mc.qis = c(mc.qis, YakoMC) ## QIS Corrected
}Iteration 10
Iteration 20
Iteration 30
Iteration 40
Iteration 50
Iteration 60
Iteration 70
Iteration 80
Iteration 90
Iteration 100 ## Trim top and bottom 2.5%
trim_ul <- function(x, p = 0.01){
x <- x[x >= quantile(x, p) & x <= quantile(x, 1-p)]
return(x)
}
cbind(mean(trim_ul(mc.qis, p = 0.025)), mean(trim_ul(mc.naive, p = 0.025)), trueZ) trueZ
[1,] 7.845875e-29 1.820323e-31 1.944557e-29Comparison
Graphically, …
library(ggplot2)
mc.data = rbind(data.frame(MC = trim_ul(mc.qis, p = 0.025), method = "QIS"),
data.frame(MC = trim_ul(mc.naive, p = 0.025), method = "Naive MC"))
(plt <- ggplot(mc.data, aes(MC, fill = method)) +
geom_histogram(alpha=0.75, bins = 30, position="identity",aes(y = after_stat(density)))+
geom_density(alpha=0.75, stat="density",position="identity",aes(y = after_stat(density)))+
expand_limits(x = c(1e-33,1e-25))+
geom_vline(xintercept=trueZ)+scale_x_log10()+
# coord_flip()+
facet_wrap(vars(method), ncol = 1, scales = "free_y")+
theme_minimal()+
labs(title = "QIS vs. Naive MC", subtitle = "Multivariate t, MVN prior"))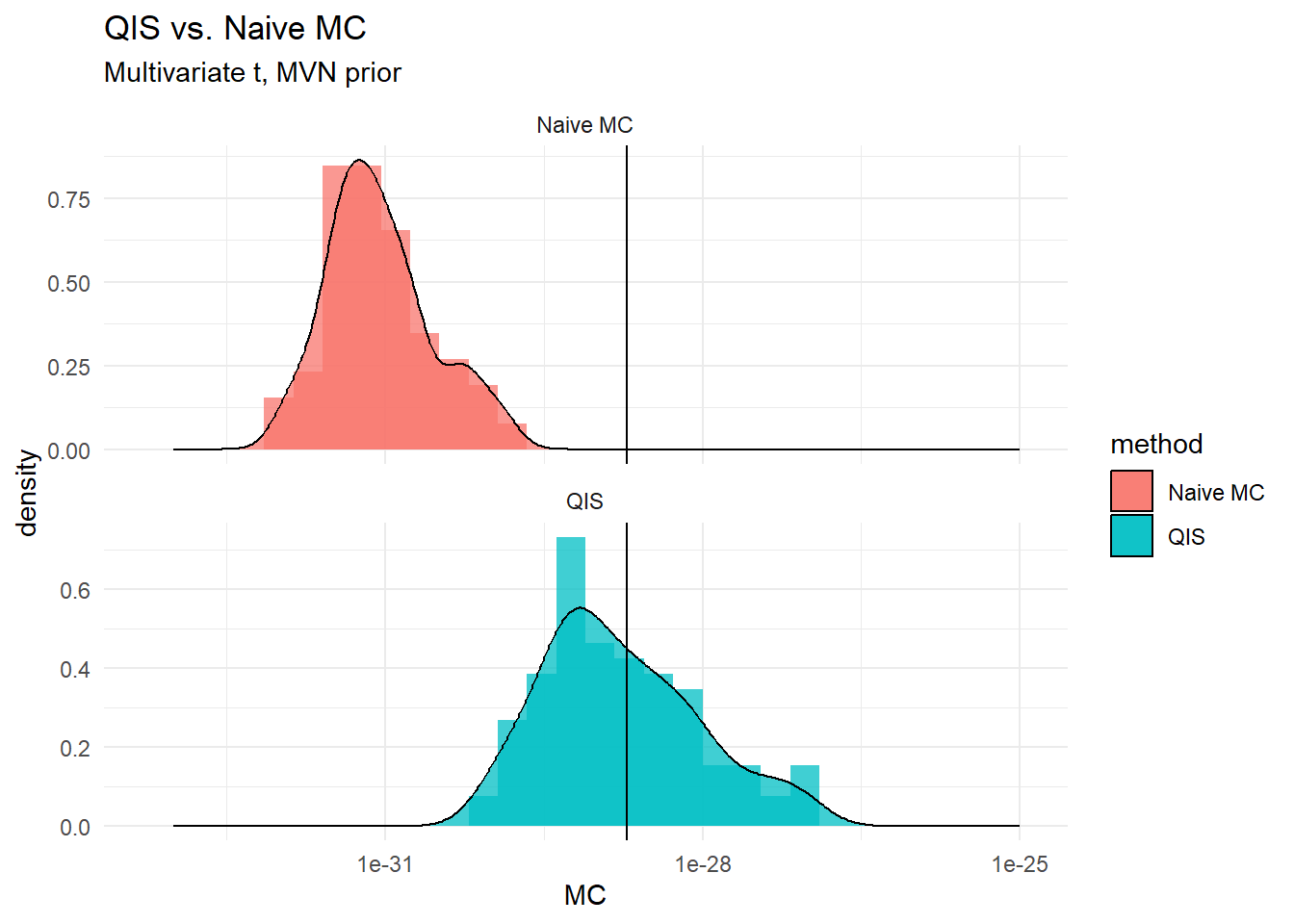
Numerically, …
## Numerical
mean.qis <- mean(mc.qis, trim = 0.025); mean.naive <- mean(mc.naive,trim = 0.025)
median.qis <- median(mc.qis); median.naive <- median(mc.naive)
mape.qis <- median(abs((mc.qis)-(trueZ))/(trueZ))
mape.naive <- median(abs(((mc.naive)-(trueZ))/(trueZ)))
rmse.qis <- sqrt((mean((mc.qis-trueZ)^2)))
rmse.naive <- sqrt((mean((mc.naive-trueZ)^2)))
perf <- rbind((cbind(mean.qis,mean.naive)),
(cbind(median.qis,median.naive)),
(cbind(mape.qis, mape.naive)),
(cbind(rmse.qis,rmse.naive)))
colnames(perf) <- c("QIS", "Naive"); row.names(perf) <- c("Mean", "Median",
"MAPE", "RMSE")
library(knitr)
library(kableExtra)
kable(perf, format = "html", digits = 99) %>%
kable_styling()| QIS | Naive | |
|---|---|---|
| Mean | 8.672069e-29 | 2.007210e-31 |
| Median | 1.376328e-29 | 7.598928e-32 |
| MAPE | 8.243520e-01 | 9.960922e-01 |
| RMSE | 3.994507e-28 | 1.913807e-29 |